Machine learning unlocks previously unseen stability
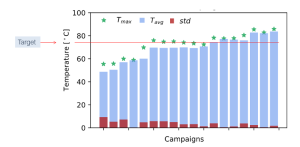
EnginZyme has an impressive track record using in silico design combined with machine learning to improve enzyme stability. In one example, we increased the melting temperature (the temperature at which a protein unfolds, and thus an implicit measure of stability) of an enzyme from 68 °C to 86 °C testing only a handful of sequences along the way. We also generated several other variants of the same enzyme that had a melting temperature greater than 80 ° C. To put these numbers into context, Francis Arnold’s group, winner of the Nobel Prize in Chemistry 2018, used a machine learning approach trained on several hundreds of previously characterised sequences to achieve a 15 °C improvement from 55 °C to 70 °C for a P450 enzyme (https://www.pnas.org/content/110/3/E193). In comparison, we achieved a greater degree of improvement with our enzyme than the Arnold group, but more importantly having screened and trained on just a fraction of the number of variants. In both cases, however, the most stable enzymes were obtained by machine learning trained on variants obtained by directed evolution and other engineering approaches. The power to access enzyme performance unseen in the training data illuminates the great potential of machine learning in enzyme engineering.
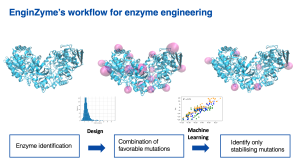
Screen – simulate – build
The in silico design involves three main steps. First, we screen enzymes that are evolutionarily related to the target enzyme for naturally occurring mutations. This helps us to identify mutations that might prove deleterious to the enzyme, as mutations that are not seen in nature are often harmful. All possible remaining mutations are then individually simulated in the target enzyme and scored in terms of their contribution to stability. Finally, the mutations that are predicted to positively contribute to stability are re-combined and introduced to the target enzyme, resulting in a handful of highly mutated variants. The stability and activity of these variants are then assessed and verified experimentally.
EnginZyme currently leverages machine learning in two ways: optimising the in silico design cycle and disentangling the effect of individual mutations on the enzyme’s stability. First, our machine learning methods use in-house data to improve how stability is scored in the in silico design cycle, resulting in a better correlation between predicted and measured stability. Second, we implement machine learning to parse out and place an uncertainty bound on the stability effect of each mutation from a group of variants with shared mutations. Using this information, we can then predict whether a different mutation to the enzyme is beneficial; a complementary and formidable way of solving the problem of enzyme stability.
Why improve enzyme stability
The standard catalysts in the chemical industry are inorganic metal catalysts, usually used in fixed bed reactors, which necessarily have a long lifetime and operate across a wide temperature range. To be competitive, enzymes must be able to compete with these properties. Increasing enzyme stability not only expands the enzyme’s useful temperature window but has been shown to correlate well with its operational lifetime- factors that have in the past prevented the widespread adoption of enzymes as the catalysts of choice. Moreover, enzyme stability also usually improves the enzyme’s tolerance towards organic solvents and reaction conditions in addition to being easier to express heterologously. In short, increased enzyme stability makes the enzyme more suitable for long term operation in a fixed bed.
Why in silico design and machine learning
Stability engineering in proteins is a difficult engineering challenge. Computational tools like in silico design and machine learning make this problem more tractable, giving us enzymes with increased thermostability faster than other methods like rational design and directed evolution. Moreover, due to our high throughput capabilities and state of the art technology, EnginZyme can produce significant volumes of high-quality data required for the successful implementation of machine learning in stability engineering. To paraphrase Moderna’s Marcello Damiani: When we run experiments, we collect even more protein stability data. This allows us to build better algorithms, which helps us build the next generation of stable proteins. It’s a virtuous cycle.
High throughput capabilities combined with state-of-the-art technology
EnginZyme’s sophisticated and effective machine learning and in silico design methods are backed up by our ability to generate a significant volume of high-quality in-house experimental data. Computational tools like machine learning can only be as good as the quality of data they are provided with. Unfortunately, a large portion of the data that is freely accessible is inconsistent as different groups often produce different results. Our high throughput capabilities, together with our state-of-the-art technology, allow us to circumvent this conundrum. We can consistently generate high-quality data for our machine learning methods. Notably, most of those pursuing machine learning on proteins are doing so on natural, or ‘wild-type’ proteins. They are not using machine learning for stability engineering, thereby providing EnginZyme with an exciting competitive advantage. Those, like EnginZyme, who are capable of generating high-quality data can continue to improve their methods, thereby increasing their competitive advantage vis a vis other competitors.
This blog post was written by Annette Egerström, Research Associate at EnginZyme. Besides improving our high-throughput methods, Annette enjoys playing the viola and drawing in her free time. We are right now looking for more researchers to join us, so if you find this post interesting, check out our open positions to find out more.
